Chemists at MIT have made a novel way to synthesize himastatin, a normal compound that has demonstrated likely as an antibiotic.
Applying their new synthesis, the researchers were being ready not only to produce himastatin but also to crank out variants of the molecule, some of which also showed antimicrobial activity. They also uncovered that the compound appears to kill microbes by disrupting their mobile membranes. The scientists now hope to structure other molecules that could have even more robust antibiotic action.
“What we want to do ideal now is master the molecular information about how it operates, so we can design and style structural motifs that could improved aid that system of motion. A good deal of our exertion proper now is to discover more about the physicochemical attributes of this molecule and how it interacts with the membrane,” suggests Mohammad Movassaghi, an MIT professor of chemistry and one of the senior authors of the examine.
Brad Pentelute, an MIT professor of chemistry, is also a senior author of the review, which seems currently in Science. MIT graduate student Kyan D’Angelo is the lead creator of the study, and graduate university student Carly Schissel is also an creator.
Mimicking character
Himastatin, which is generated by a species of soil micro organism, was initially uncovered in the 1990s. In animal studies, it was located to have anticancer action, but the needed doses experienced poisonous aspect results. The compound also showed possible antimicrobial action, but that probable has not been explored in element, Movassaghi says.
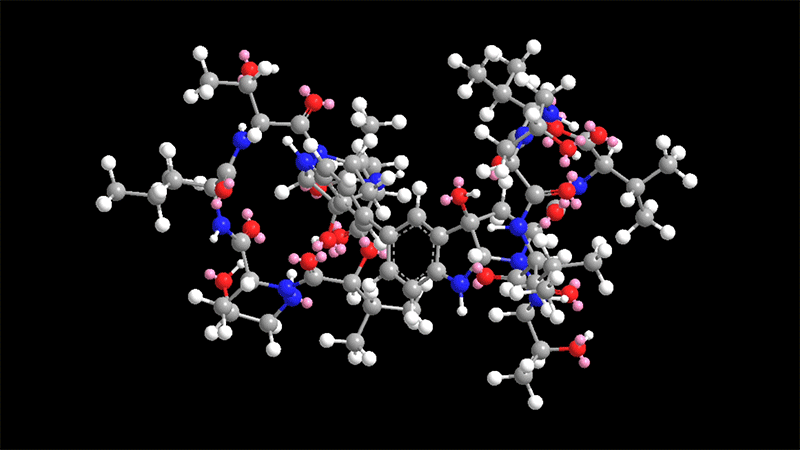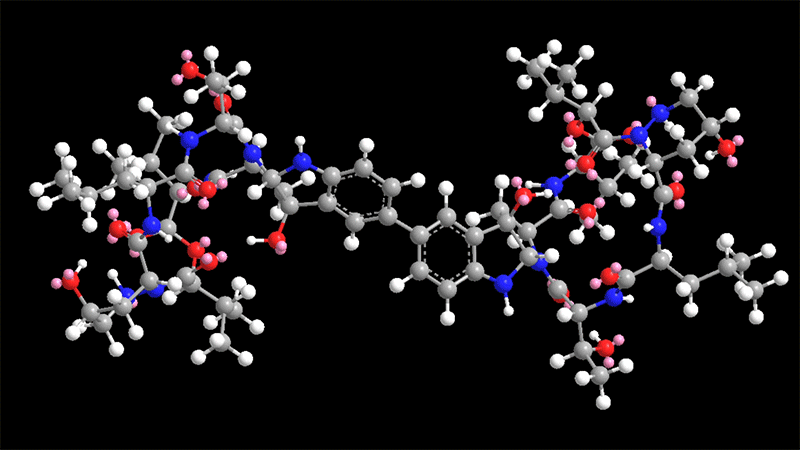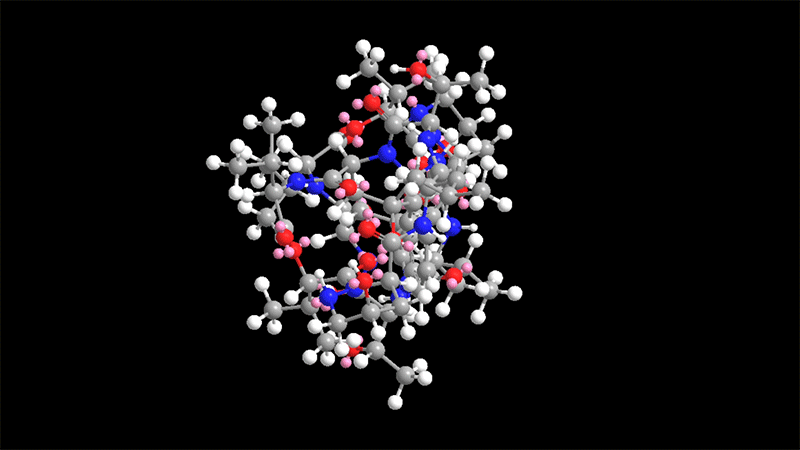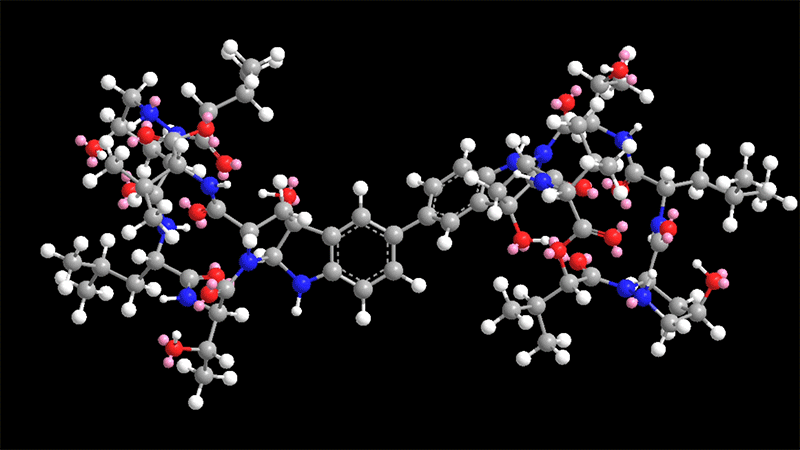
Himastatin is a elaborate molecule that is made up of two identical subunits, known as monomers, that be a part of jointly to sort a dimer. The two subunits are hooked with each other by a bond that join a 6-carbon ring in just one of the monomers to the equivalent ring in the other monomer.
This carbon-carbon bond is significant for the molecule’s antimicrobial activity. In past initiatives to synthesize himastatin, researchers have tried using to make that bond to start with, working with two uncomplicated subunits, and then additional extra elaborate chemical teams on to the monomers.
The MIT team took a unique strategy, motivated by the way this response is executed in bacteria that create himastatin. Those bacteria have an enzyme that can sign up for the two monomers as the extremely final move of the synthesis, by turning every single of the carbon atoms that will need to be joined together into extremely reactive radicals.
To mimic that system, the researchers to start with developed elaborate monomers from amino acid creating blocks, assisted by a quick peptide synthesis technologies beforehand created by Pentelute’s lab.
“By making use of stable-phase peptide synthesis, we could rapidly-forward by means of quite a few synthetic measures and mix-and-match building blocks quickly,” D’Angelo claims. “That’s just one of the ways that our collaboration with the Pentelute Lab was incredibly practical.”
The scientists then employed a new dimerization technique made in the Movassaghi lab to hook up two complex molecules together. This new dimerization is dependent on the oxidation of aniline to variety carbon radicals in each and every molecule. These radicals can respond to form the carbon-carbon bond that hooks the two monomers with each other. Utilizing this strategy, the scientists can generate dimers that comprise distinctive types of subunits, in addition to the natural way transpiring himastatin dimers.
“The reason we received energized about this form of dimerization is for the reason that it enables you to seriously diversify the construction and accessibility other possible derivatives very speedily,” Movassaghi says.
Membrane disruption
A single of the variants that the researchers made has a fluorescent tag, which they utilised to visualize how himastatin interacts with bacterial cells. Working with these fluorescent probes, the researchers uncovered that the drug accumulates in the bacterial cell membranes. This led them to hypothesize that it will work by disrupting the cell membrane, which is also a mechanism utilised by at minimum a single Food and drug administration-accepted antibiotic, daptomycin.
The scientists also developed various other himastatin variants by swapping in unique atoms in unique parts of the molecule, and examined their antimicrobial action towards six bacterial strains. They observed that some of these compounds experienced strong action, but only if they provided just one obviously developing monomer along with 1 that was various.
“By bringing two total halves of the molecule with each other, we could make a himastatin spinoff with only a solitary fluorescent label. Only with this edition could we do microscopy scientific tests that presented proof of himastatin’s localization in just bacterial membranes, because symmetric versions with two labels did not have the proper exercise,” D’Angelo suggests.
Andrew Myers, a professor of chemistry at Harvard College, says that the new synthesis functions “fascinating new chemical improvements.”
“This strategy permits oxidative dimerization of absolutely synthetic monomer subunits to put together the antibiotic himastatin, in a method similar to its biosynthesis,” says Myers, who was not concerned in the exploration. “By synthesizing a range of analogs, critical framework-exercise relationships were being unveiled, as well as proof that the organic merchandise capabilities at the amount of the bacterial envelope.”
The researchers now plan to design a lot more variants that they hope might have extra strong antibiotic exercise.
“We’ve already recognized positions that we can derivatize that could potentially either keep or improve the action. What is definitely thrilling to us is that a important selection of the derivatives that we accessed by this design and style course of action retain their antimicrobial action,” Movassaghi says.
The investigate was funded by the Nationwide Institutes of Overall health, the All-natural Sciences and Engineering Investigation Council of Canada, and a Nationwide Science Basis graduate analysis fellowship.
